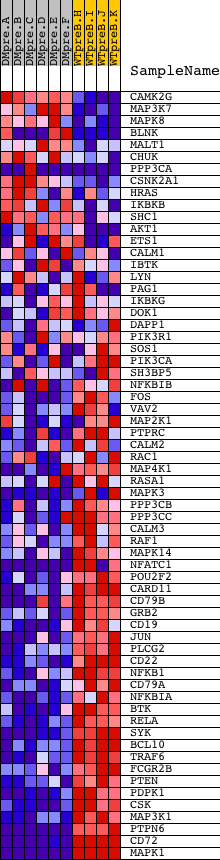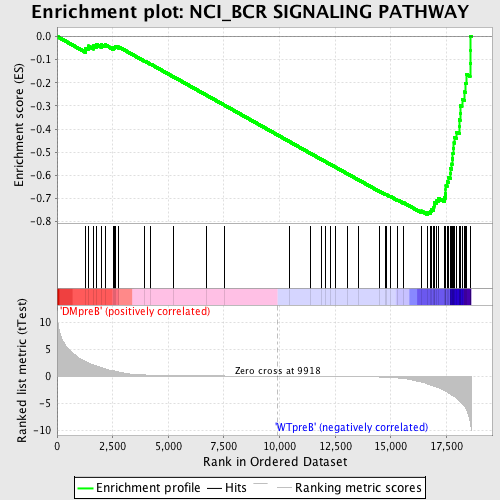
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | NCI_BCR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.7689641 |
| Normalized Enrichment Score (NES) | -1.7866076 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.002428016 |
| FWER p-Value | 0.0080 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CAMK2G | 21905 | 1270 | 2.700 | -0.0505 | No | ||
| 2 | MAP3K7 | 16255 | 1398 | 2.420 | -0.0413 | No | ||
| 3 | MAPK8 | 6459 | 1644 | 2.061 | -0.0408 | No | ||
| 4 | BLNK | 23681 3691 | 1758 | 1.877 | -0.0344 | No | ||
| 5 | MALT1 | 6274 | 1985 | 1.573 | -0.0361 | No | ||
| 6 | CHUK | 23665 | 2157 | 1.372 | -0.0362 | No | ||
| 7 | PPP3CA | 1863 5284 | 2520 | 0.993 | -0.0491 | No | ||
| 8 | CSNK2A1 | 14797 | 2578 | 0.947 | -0.0459 | No | ||
| 9 | HRAS | 4868 | 2610 | 0.906 | -0.0415 | No | ||
| 10 | IKBKB | 4907 | 2775 | 0.746 | -0.0454 | No | ||
| 11 | SHC1 | 9813 9812 5430 | 3936 | 0.225 | -0.1064 | No | ||
| 12 | AKT1 | 8568 | 4218 | 0.183 | -0.1204 | No | ||
| 13 | ETS1 | 10715 6230 3135 | 5251 | 0.099 | -0.1753 | No | ||
| 14 | CALM1 | 21184 | 6697 | 0.049 | -0.2529 | No | ||
| 15 | IBTK | 19047 | 7506 | 0.033 | -0.2962 | No | ||
| 16 | LYN | 16281 | 10463 | -0.007 | -0.4555 | No | ||
| 17 | PAG1 | 15376 | 11377 | -0.020 | -0.5046 | No | ||
| 18 | IKBKG | 2570 2562 4908 | 11885 | -0.028 | -0.5317 | No | ||
| 19 | DOK1 | 17104 1018 1177 | 12045 | -0.031 | -0.5401 | No | ||
| 20 | DAPP1 | 11159 6445 6446 | 12268 | -0.036 | -0.5518 | No | ||
| 21 | PIK3R1 | 3170 | 12502 | -0.041 | -0.5641 | No | ||
| 22 | SOS1 | 5476 | 13046 | -0.056 | -0.5930 | No | ||
| 23 | PIK3CA | 9562 | 13540 | -0.075 | -0.6191 | No | ||
| 24 | SH3BP5 | 6278 | 14484 | -0.150 | -0.6689 | No | ||
| 25 | NFKBIB | 17906 | 14778 | -0.198 | -0.6834 | No | ||
| 26 | FOS | 21202 | 14800 | -0.201 | -0.6832 | No | ||
| 27 | VAV2 | 5848 2670 | 14978 | -0.236 | -0.6911 | No | ||
| 28 | MAP2K1 | 19082 | 15318 | -0.325 | -0.7072 | No | ||
| 29 | PTPRC | 5327 9662 | 15564 | -0.428 | -0.7176 | No | ||
| 30 | CALM2 | 8681 | 16368 | -1.071 | -0.7538 | No | ||
| 31 | RAC1 | 16302 | 16651 | -1.434 | -0.7594 | Yes | ||
| 32 | MAP4K1 | 18313 | 16775 | -1.624 | -0.7553 | Yes | ||
| 33 | RASA1 | 10174 | 16815 | -1.689 | -0.7461 | Yes | ||
| 34 | MAPK3 | 6458 11170 | 16931 | -1.809 | -0.7403 | Yes | ||
| 35 | PPP3CB | 5285 | 16941 | -1.823 | -0.7287 | Yes | ||
| 36 | PPP3CC | 21763 | 16961 | -1.850 | -0.7174 | Yes | ||
| 37 | CALM3 | 8682 | 17069 | -2.011 | -0.7098 | Yes | ||
| 38 | RAF1 | 17035 | 17131 | -2.125 | -0.6989 | Yes | ||
| 39 | MAPK14 | 23313 | 17421 | -2.682 | -0.6967 | Yes | ||
| 40 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 17449 | -2.738 | -0.6799 | Yes | ||
| 41 | POU2F2 | 17929 | 17474 | -2.802 | -0.6626 | Yes | ||
| 42 | CARD11 | 8436 | 17478 | -2.819 | -0.6440 | Yes | ||
| 43 | CD79B | 20185 1309 | 17530 | -2.928 | -0.6273 | Yes | ||
| 44 | GRB2 | 20149 | 17590 | -3.055 | -0.6102 | Yes | ||
| 45 | CD19 | 17640 | 17676 | -3.314 | -0.5927 | Yes | ||
| 46 | JUN | 15832 | 17687 | -3.335 | -0.5711 | Yes | ||
| 47 | PLCG2 | 18453 | 17743 | -3.480 | -0.5509 | Yes | ||
| 48 | CD22 | 17881 | 17755 | -3.507 | -0.5282 | Yes | ||
| 49 | NFKB1 | 15160 | 17789 | -3.591 | -0.5061 | Yes | ||
| 50 | CD79A | 18342 | 17804 | -3.629 | -0.4827 | Yes | ||
| 51 | NFKBIA | 21065 | 17838 | -3.699 | -0.4599 | Yes | ||
| 52 | BTK | 24061 | 17851 | -3.733 | -0.4357 | Yes | ||
| 53 | RELA | 23783 | 17959 | -4.092 | -0.4143 | Yes | ||
| 54 | SYK | 21636 | 18074 | -4.548 | -0.3902 | Yes | ||
| 55 | BCL10 | 15397 | 18081 | -4.569 | -0.3601 | Yes | ||
| 56 | TRAF6 | 5797 14940 | 18115 | -4.665 | -0.3309 | Yes | ||
| 57 | FCGR2B | 8959 | 18125 | -4.712 | -0.3000 | Yes | ||
| 58 | PTEN | 5305 | 18200 | -4.996 | -0.2708 | Yes | ||
| 59 | PDPK1 | 23097 | 18324 | -5.661 | -0.2398 | Yes | ||
| 60 | CSK | 8805 | 18359 | -5.871 | -0.2026 | Yes | ||
| 61 | MAP3K1 | 21348 | 18411 | -6.282 | -0.1636 | Yes | ||
| 62 | PTPN6 | 17002 | 18561 | -8.216 | -0.1170 | Yes | ||
| 63 | CD72 | 8718 | 18585 | -8.914 | -0.0589 | Yes | ||
| 64 | MAPK1 | 1642 11167 | 18592 | -9.104 | 0.0013 | Yes |